Comparing with SOTA Methods
To further thoroughly evaluate the performance of DeepSeq2drug, we conducted some extra experiments. We first compared GCNMDA [1], which is built on the Drug Virus Info 1 database. For Drug Virus Info version 1 [2], the viral names are noted with the virus name. Nonetheless, in Drug Virus Info version 2 [3], the viral names are noted with abbreviations(virus name also provided, however slightly different from version 1).
Thus, we compared our methods with corresponding SOTA methods based on different database versions. (Part I, for drugvirus infor 1, Part II, for drug virus infor 2).
First of all, we compared the SOTA method using statistics features. The results are shown in Table 1. Compared with SOTA methods, our methods are multi-modal and can be easily expanded. Also, we provided embeddings, GitHub repos, websites, and software for better reproducibility.
| Method | Type | E/G/W/S/N | Expand | Drug | Virus | Pairs | Known pairs |
| GCNMDA [1] | GB | 11000 | No | 175 | 95 | 933 | Drug virus info |
| AntiViralDL [4] | GB | 01000 | No | 336 | 158 | 1,648 | Drug virus info 2+ manually collected |
| DeepSeq2Drug | MM | 11111 | Yes | 9212 + | 103 + | 1156 | Drug virus info 2 |
| Increment (compared with GCNMDA) | – | – | 5164% | 9.5% | 24% | – |
Table 1. Comparison with SOTA methods in statistic
*GB denotes graph-based (Network-based) methods, and MM denotes multi-modal methods. Embedding means providing embedding for viruses and drugs. E/G/W/S/N refers to whether the methods provide embedding/ GitHub repo/websites or webserver/software/novel virus solution. The number ‘1’ means provided by authors, while ‘0’ means not provided. The number series ‘11111‘ means that all of these elements are equipped/provided. The symbol + means that the number can be added easily.
Part I compares the SOTA method using Drug virus info 1
We designed experiments using GCNMDA’s feature as input and tested them in our framework.
That database contains some overlaps (such as in version one, it contains four viruses named: Middle East respiratory syndrome coronavirus, Middle East respiratory tract syndrome coronavirus, Middle East resporitary syndrome coronavirus, Middle Eastern respiratory syndrome virus. However, the second version (Drug Virus info 2) only contains the Middle East respiratory syndrome-related coronavirus. In this situation, we averaged those four embedding together and labeled the results embedding as Middle East respiratory syndrome-related coronavirus. After averaging GCNMDA and our viral embeddings, we conducted the experiments on the constructed dataset, in which the negative samples were generally limited inside the GCNMDA’s virus and drug list to compare it fairly. We implemented five-fold cross-validation, repeated it five times, and contacted all the valid results to generate the ROC/PR curves. For vdkeys, we selected the nine vdkeys that the feature pool selected. The results are shown below. As can be seen from the figure below(Figure1, Figure2), almost all single models for selected vdkey pairs can outperform averaged GCNMDA embedding in both ROC and PR curves.
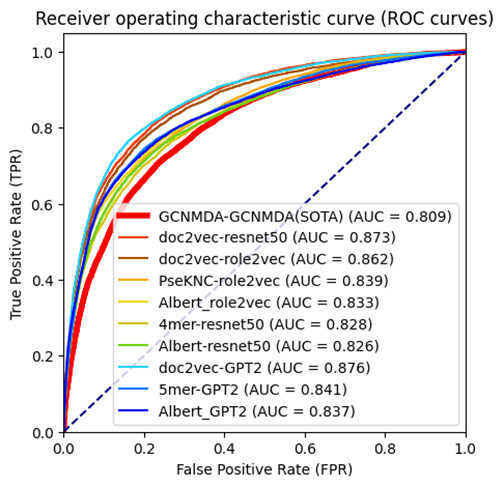
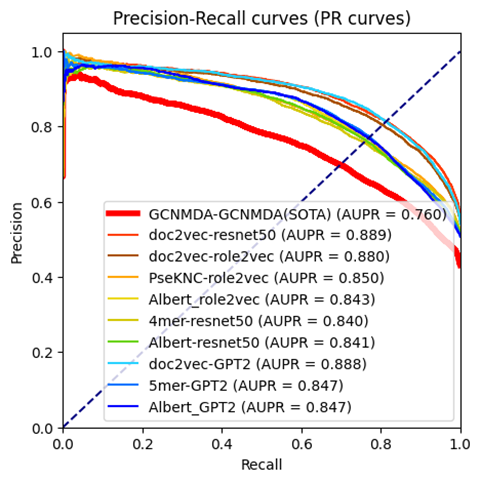
Figure1, Figure2, ROC/PR curves of different vdkeys using the dataset of Drugvirus info 1.
Part II compares the SOTA method using Drug virus info 2.
To further test the prediction ability of the proposed method, we found another recently published paper, AntiViralDL[4], which was published in another top journal, IEEE JBHI, on 03 November 2023. They provided code and dataset but didn’t provide embedding for each virus and drug. We implemented their codes and tried to generate viral/drug embedding by extracting 1318 embedding of virus-drug pairs. After filtering with our database (Virus/ Drug database), we finally got 264 drug embeddings (with DBID) and 151 viral embeddings (with Virus abbreviations). We named viral embeddings from AntiViralDL as ‘anti’ and drug embeddings as ‘anti.’ Thus, the vdkey for this method is noted as ‘Anti-Anti’.
To align with AntiViralDL, we replaced the virus name with Abbreviation and conducted the experiments under two conditions. Note that AntiViralDL is also a graph-based method. Thus, we combined their graph-based feature (viral and drug embedding) with our expandable drug features (resnet50, biogpt, role2vec). As different negative sampling methods might contribute to different results, we implemented the comparison under two conditions:
- Condition one:
We leveraged the anti-virals training dataset (positive samples and negative samples on the training set and conducted experiments using our embeddings). They provided five training datasets in their GitHub repo. Thus, we conducted five-fold cross-validation on these five training datasets and then calculated the AUC/AUPR values based on those datasets.
- Condition two
We leveraged our method to generate positive samples and negative samples and compared anti-virals’ viral and drug embedding in our framework. We repeated five-fold cross-validation five times with varied dividing random seeds.
Results
We analyzed and compared results in three ways. The first one is a table indicating the average AUC/AUPR/metric, and the second one is a Violin-Swam figure indicating the distribution of
, the third one is ROC/PR curves of all valid results indicating the overall prediction performances.
Condition one:
The average AUC/AUPR/metric values of the mix-embedding combinations are shown in Table 2, with the condition of using the AntiViralDL dataset. From the Table, the doc2vec-based viral sequence embedding combination can outperform most of the expandable drug embeddings (GPT-based, resnet-based drug embeddings). From Figure 3, Anti_Anti vdkey can significantly outperform, Anti_3Dresne50, Anti_bioGPT, Anti_role2vec, while but not significantly better than doc2vec-based vdkeys. From Figure 4, Doc2vec_bioGPT vdkey can outperform most of the Anti-participated-embedding combinations, except for Anti_Anti. It has the same trend in the metric, except that Doc2vec_bioGPT is not significantly better than Doc2vec_Anti. This is reasonable, as the Anti-embeddings (both viral and drug) are trained based on the known database ( the graph-based methods need known pairs to construct the graph or network). Also, the results show that when expandable drug embeddings (GPT-based, resnet-based drug embeddings) were introduced to Anti-viral embedding, the AUC/AUPR decreased rapidly. This indicates that network-based viral embedding might not be suitable for the concatenation dataset.
Table 2. Comparison with AntiViralDL [4] Embedding leverage AntiViralDL dataset
| Embeddings(vdkeys) | AUC | AUC_std | AUPR | AUPR_std | Metric |
| Anti_3Dresne50 | 0.7898 | 0.0209 | 0.7963 | 0.0297 | 1.586 |
| Anti_Anti | 0.8493 | 0.0153 | 0.8351 | 0.03 | 1.6844 |
| Anti_bioGPT | 0.8141 | 0.0171 | 0.8094 | 0.0242 | 1.6235 |
| Anti_role2vec | 0.8187 | 0.0184 | 0.8227 | 0.0312 | 1.6414 |
| Doc2vec_3Dresnet50 | 0.8419 | 0.0444 | 0.8533 | 0.0439 | 1.6952 |
| Doc2vec_Anti | 0.834 | 0.0331 | 0.8209 | 0.0421 | 1.6549 |
| Doc2vec_bioGPT | 0.8468 | 0.0239 | 0.8575 | 0.0281 | 1.7044 |
| Doc2vec_role2vec | 0.8427 | 0.037 | 0.8549 | 0.0413 | 1.6976 |
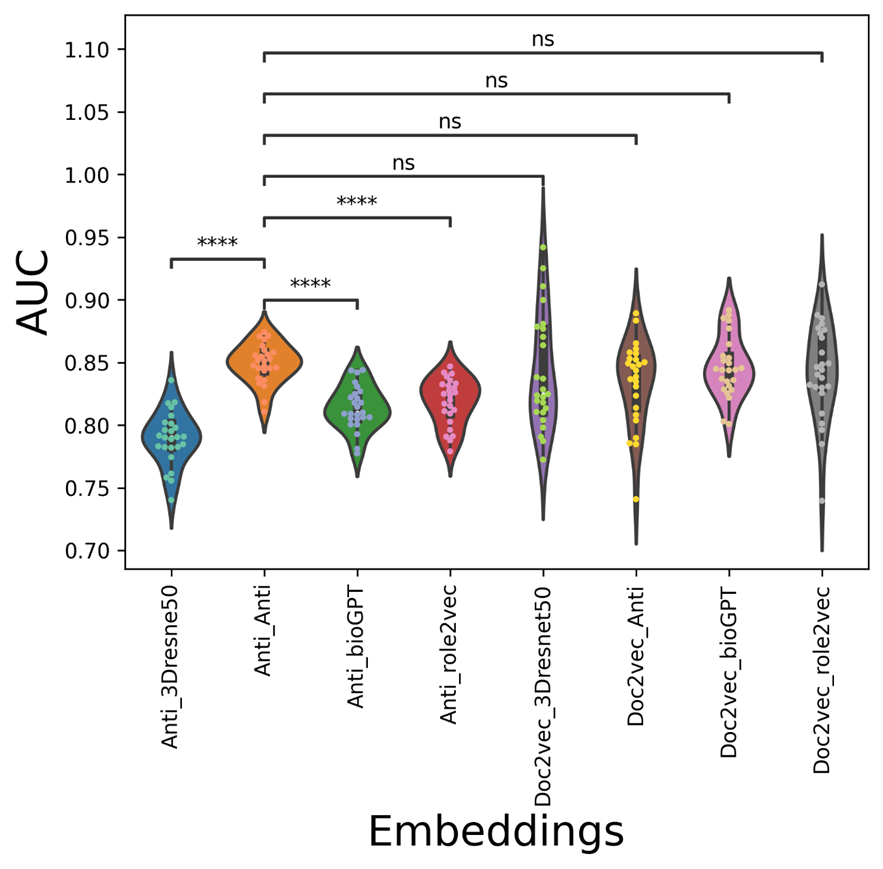
Figure 3. The AUCviolin-swam figure of different vdkeys in condition one: leveraging AntiViralDL dataset.
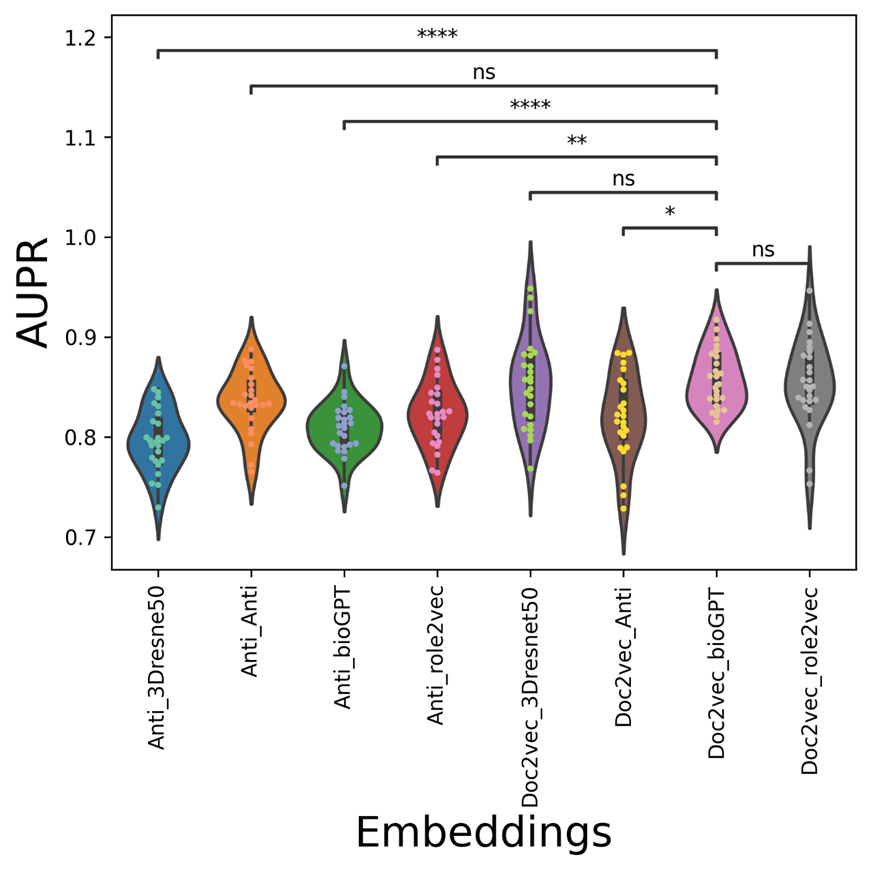
Figure 4. The AUPR violin-swam figure of different vdkeys in condition one: leveraging AntiViralDL dataset.
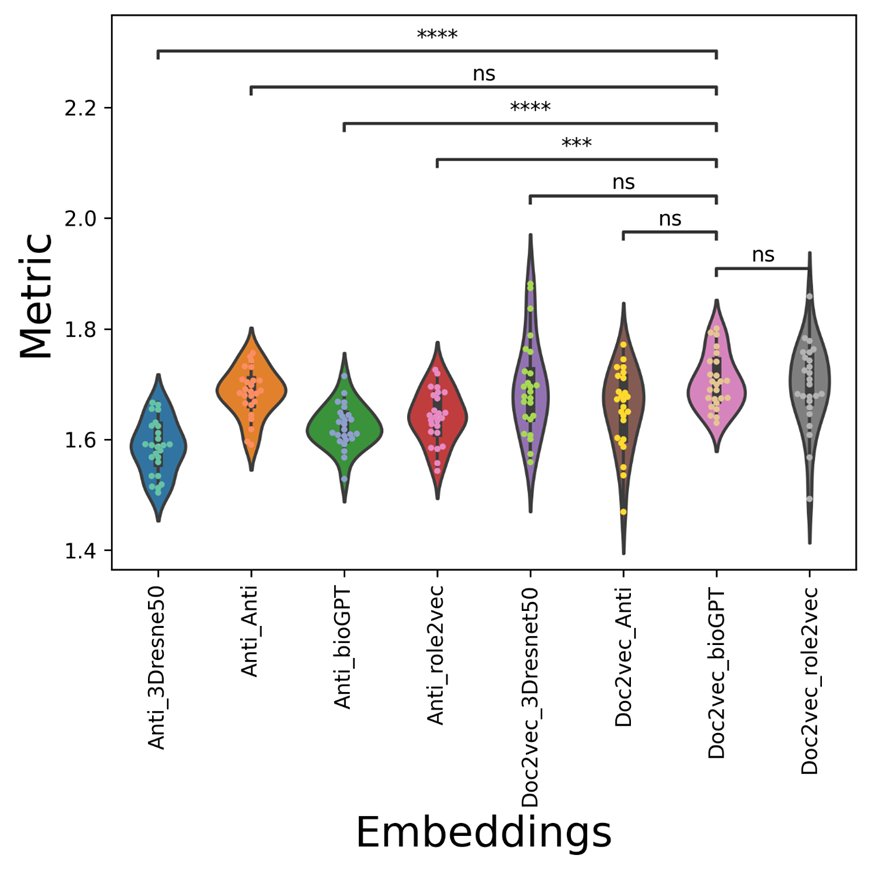
Figure 5. The metric violin-swam figure of different vdkeys in condition one: leveraging AntiViralDL dataset.
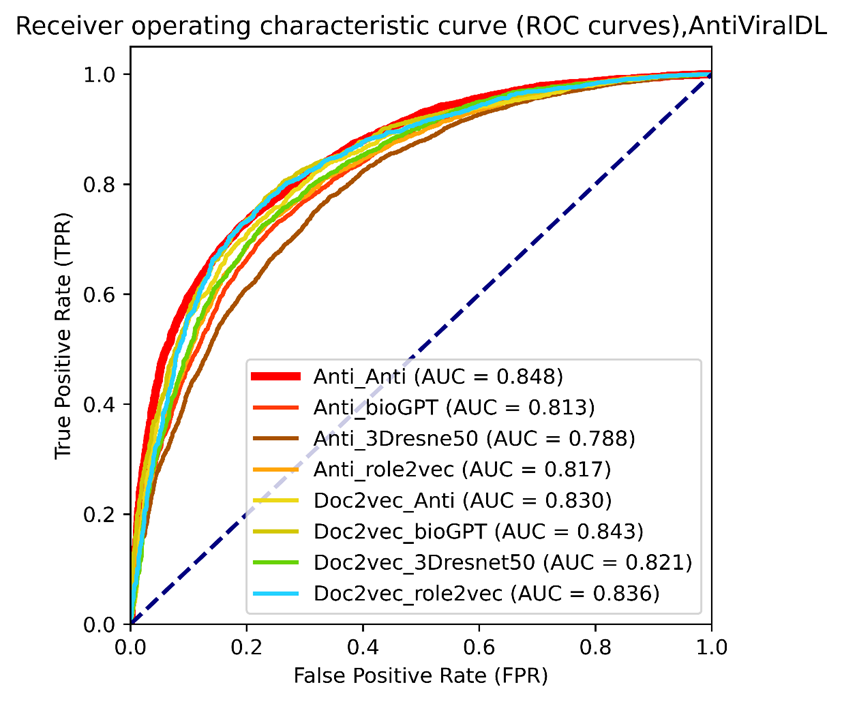
Figure 6. The ROC curves of selected vdkeys were calculated using the AntiViralDL training dataset
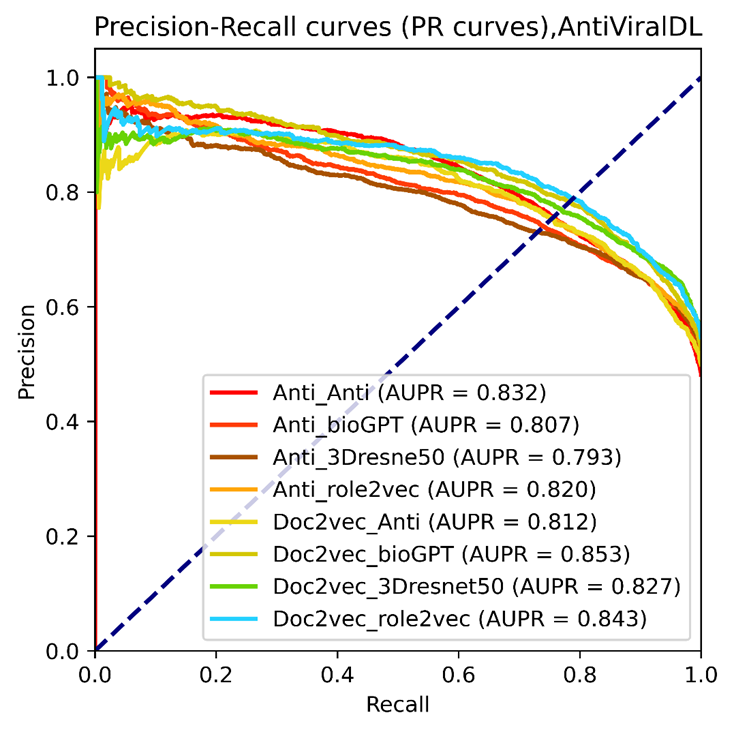
Figure 7. The PR curves of selected vdkeys were calculated using the AntiViralDL training dataset
Condition two:
The average AUC/AUPR/metric values of the mix-embedding combinations are shown in Table 3, with the condition of using Our drugvirus dataset. From the point of view of the average metric value, the AUC values of doc2vec-related vdkey (except Doc2vec_Anti) are higher than Anti-related vdkeys in both AUC and metric. From Figure 8, The Doc2vec_bioGPT vdkey now outperforms all anti-participated vdkeys significantly. As can be seen from Figure 9, although anti_3Dresnet50 can get a higher mean AUPR with the value of 0.8185( which equals Doc2vec_bioGPT), it is only significantly better than Anti_Anti and Doc2vec_Anti. From Figure 10, the distribution of the metric indicates that the Doc2vec_bioGPT of the metric is significantly higher than the rest of the anti-participated-embedding combinations.
Compared with condition one, the Doc2vec_bioGPT in condition two compared with Anti_Anti and Doc2vev_Anti becomes significantly better/higher. This might be possible as graph-based methods are usually limited in the number of drugs.
Table 3. Comparison with AntiViralDL [4] Embedding leverage Our dataset
| Embeddings(vdkeys) | AUC | AUC_std | AUPR | AUPR_std | Metric |
| Anti_3Dresne50 | 0.8139 | 0.0217 | 0.8185 | 0.0281 | 1.6324 |
| Anti_Anti | 0.8005 | 0.0194 | 0.7956 | 0.0253 | 1.5961 |
| Anti_bioGPT | 0.8103 | 0.0215 | 0.8078 | 0.0294 | 1.6181 |
| Anti_role2vec | 0.8169 | 0.0235 | 0.8118 | 0.0312 | 1.6288 |
| Doc2vec_3Dresnet50 | 0.8506 | 0.028 | 0.8126 | 0.0409 | 1.6632 |
| Doc2vec_Anti | 0.8069 | 0.0244 | 0.7651 | 0.0378 | 1.572 |
| Doc2vec_bioGPT | 0.858 | 0.0235 | 0.8185 | 0.0354 | 1.6765 |
| Doc2vec_role2vec | 0.8482 | 0.0238 | 0.7969 | 0.0372 | 1.6451 |
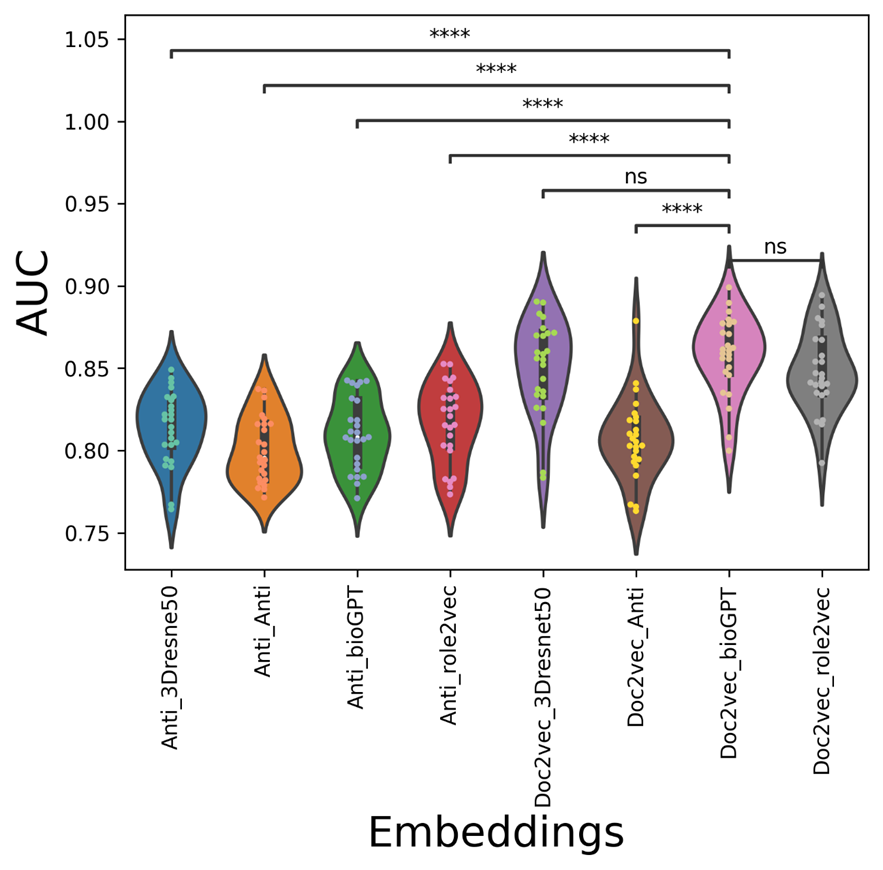
Figure 8. The AUC violin-swam figure of different vdkeys in condition one: leveraging our drugvirus dataset.
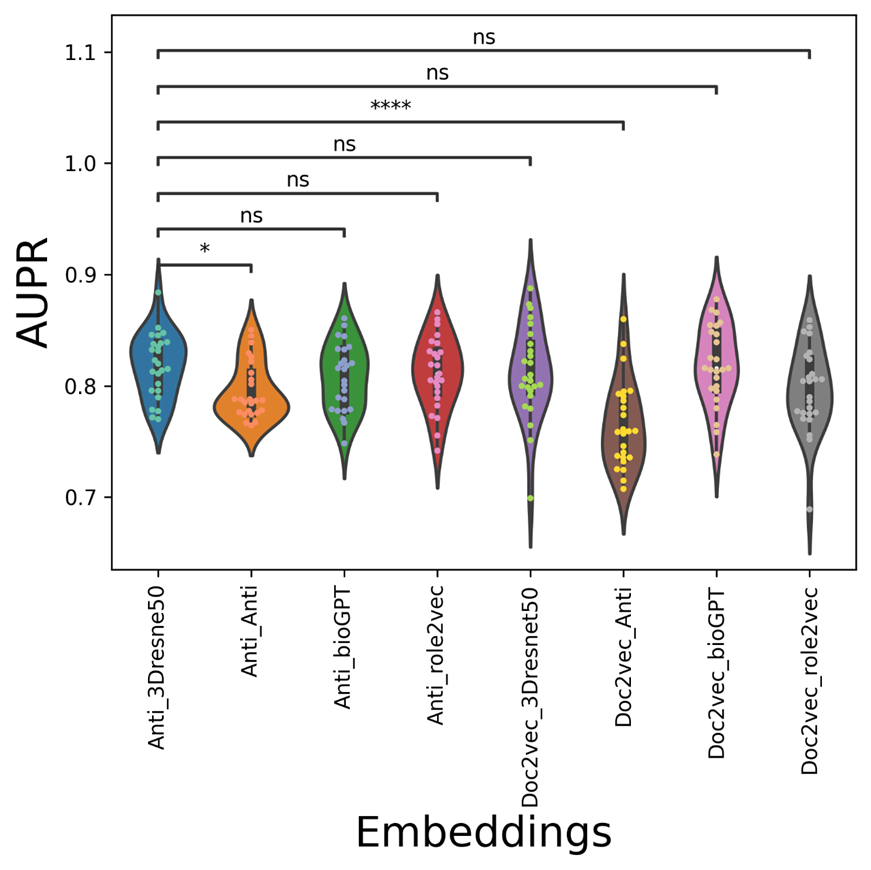
Figure 9. The AUPR violin-swam figure of different vdkeys in condition one: leveraging our drugvirus dataset.
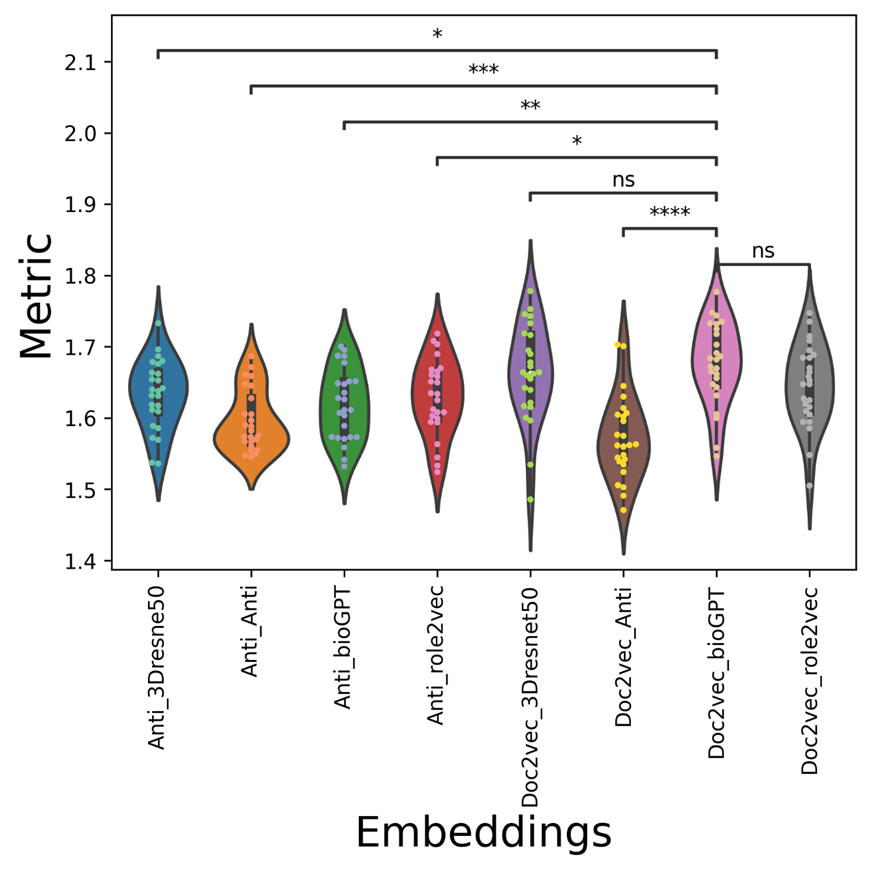
Figure 10. The metric violin-swam figure of different vdkeys in condition one: leveraging our drugvirus dataset.
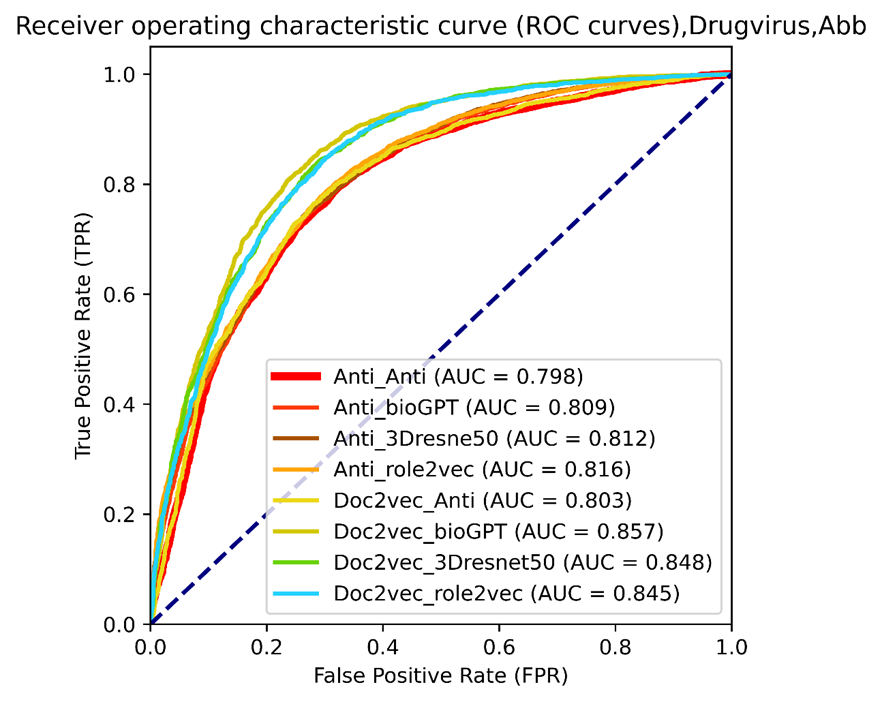
Figure 11. The ROC curves of selected vdkeys were calculated using our drug virus training dataset
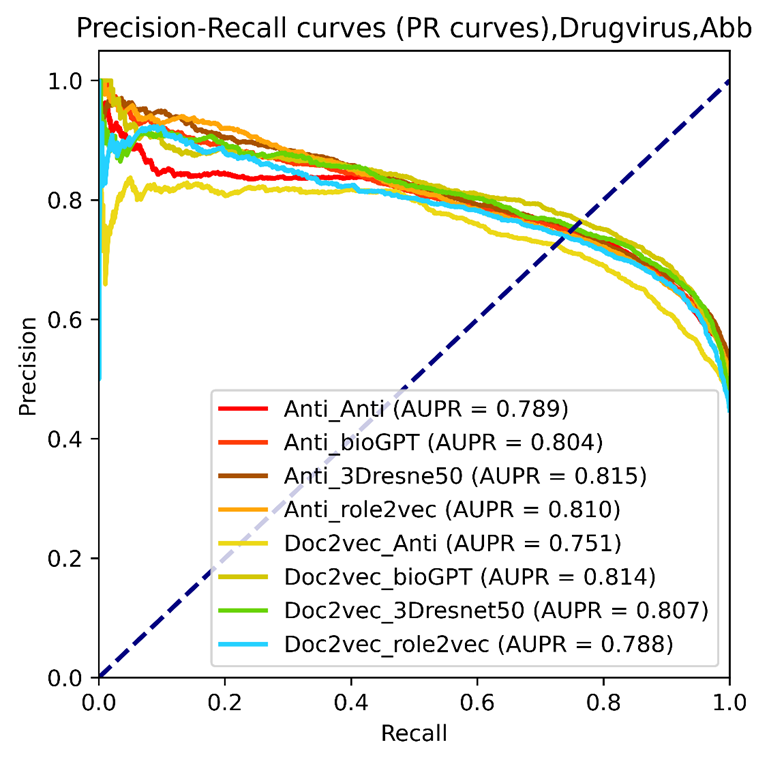
Figure 12. The PR curves of selected vdkeys were calculated using our drug virus training dataset
Summary:
In Drug Virus Info 1 [2], when averaging the GCNMDA embedding and re-conducting the experiments, we find that any of the selected nine vdkey pairs are better than GCNMDA-GCNMDA vdkey combinations.
In the Drug Virus Info 2 [3] datasets, under condition one, the graph-based methodsAntiViralDL have a relatively higher mean AUC. However, it is not significantly higher than Doc2vec-related vdkeys. Furthermore, by replacing the AntiViralDL training dataset with our drug virus dataset, the results of the highest doc2vec-related vdkey become significantly higher than Anti-related vdkeys.
Secondly, the number of graph-based drug embeddings is limited; by replacing the anti-drug embedding with expandable embedding from other feature domains (modalities), the performance could decrease (under the first condition), which indicates that expanding the graph-based methods, such as GCNMDA or AntiViralDL, would not be suitable for contacting them directly with other expandable embeddings. The trend seems different in the second condition; maybe it results from the limited number of anti-drug embedding contains.
Anyway, no matter the condition of one or two, the doc2vec-related embedding vdkeys can have a higher mean and significantly higher metric value than most anti-participated embedding vdkeys, which indicates the potential better-predicted ability of proposed embedding generation methods. (at least in the situation of feature concatenation.)
- Long Y, Wu M, Kwoh C K, et al. Predicting human microbe–drug associations via graph convolutional network with conditional random field[J]. Bioinformatics, 2020, 36(19): 4918-4927.
- Andersen P I, Ianevski A, Lysvand H, et al. Discovery and development of safe-in-man broad-spectrum anti-viral agents[J]. International Journal of Infectious Diseases, 2020, 93: 268-276.
- Ianevski A, Simonsen R M, Myhre V, et al. DrugVirus. info 2.0: an integrative data portal for broad-spectrum anti-virals (BSA) and BSA-containing drug combinations (BCCs)[J]. Nucleic Acids Research, 2022, 50(W1): W272-W275.
- Zhang P, Hu X, Li G, et al. AntiViralDL: Computational Anti-viral Drug Repurposing Using Graph Neural Network and Self-Supervised Learning[J]. IEEE Journal of Biome